Density, distribution function, quantile function, and random
generation for the zero-hurdle negative binomial distribution with
parameters mu, theta (or size), and pi.
Usage
dhnbinom(x, mu, theta, size, pi, log = FALSE)
phnbinom(q, mu, theta, size, pi, lower.tail = TRUE, log.p = FALSE)
qhnbinom(p, mu, theta, size, pi, lower.tail = TRUE, log.p = FALSE)
rhnbinom(n, mu, theta, size, pi)Arguments
- x
vector of (non-negative integer) quantiles.
- mu
vector of (non-negative) negative binomial location parameters.
- theta, size
vector of (non-negative) negative binomial overdispersion parameters. Only
thetaor, equivalently,sizemay be specified.- pi
vector of zero-hurdle probabilities in the unit interval.
- log, log.p
logical indicating whether probabilities p are given as log(p).
- q
vector of quantiles.
- lower.tail
logical indicating whether probabilities are \(P[X \le x]\) (lower tail) or \(P[X > x]\) (upper tail).
- p
vector of probabilities.
- n
number of random values to return.
Details
All functions follow the usual conventions of d/p/q/r functions
in base R. In particular, all four hnbinom functions for the
hurdle negative binomial distribution call the corresponding nbinom
functions for the negative binomial distribution from base R internally.
Note, however, that the precision of qhnbinom for very large
probabilities (close to 1) is limited because the probabilities
are internally handled in levels and not in logs (even if log.p = TRUE).
Examples
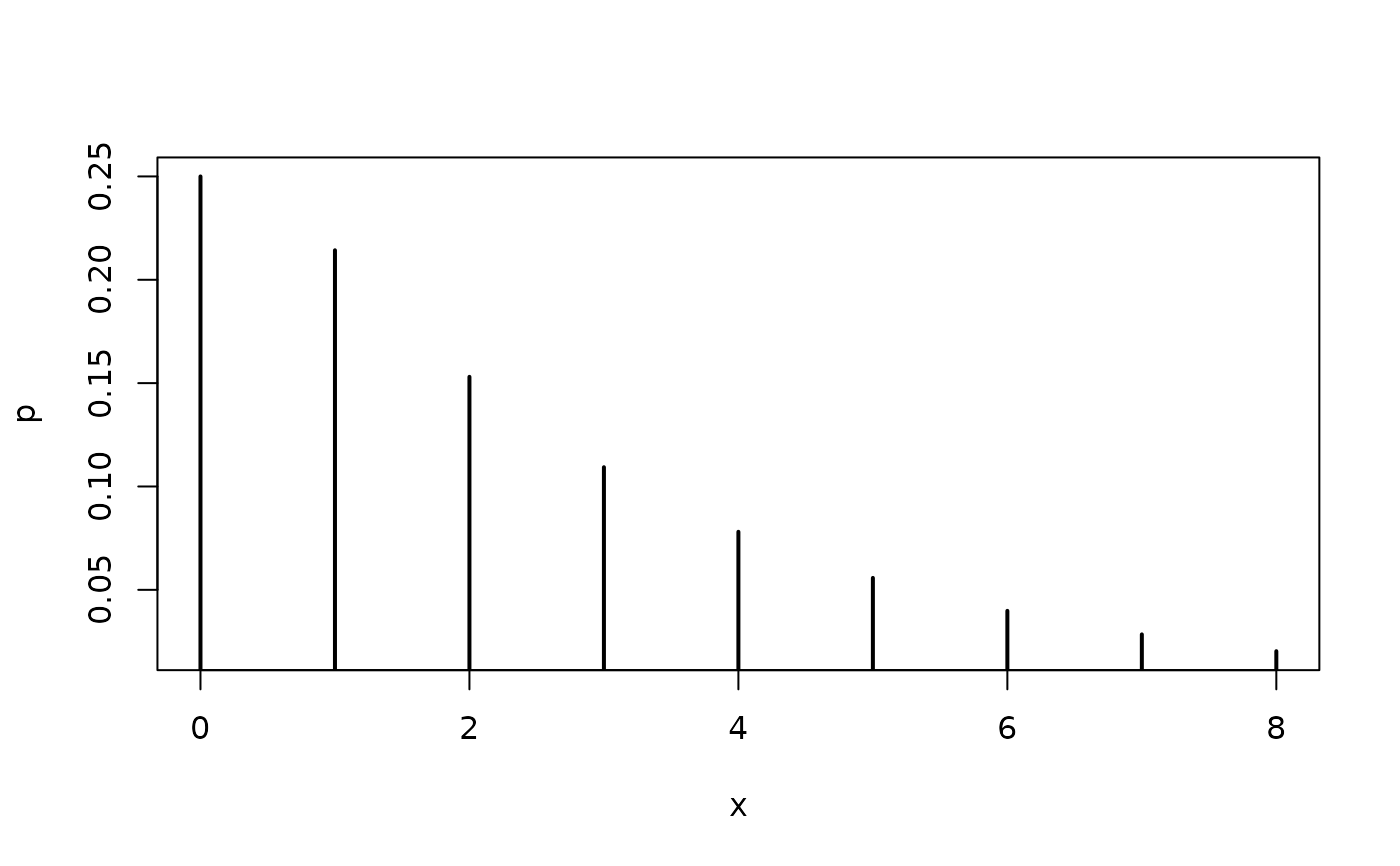
## theoretical probabilities for a hurdle negative binomial distribution
x <- 0:8
p <- dhnbinom(x, mu = 2.5, theta = 1, pi = 0.75)
plot(x, p, type = "h", lwd = 2)
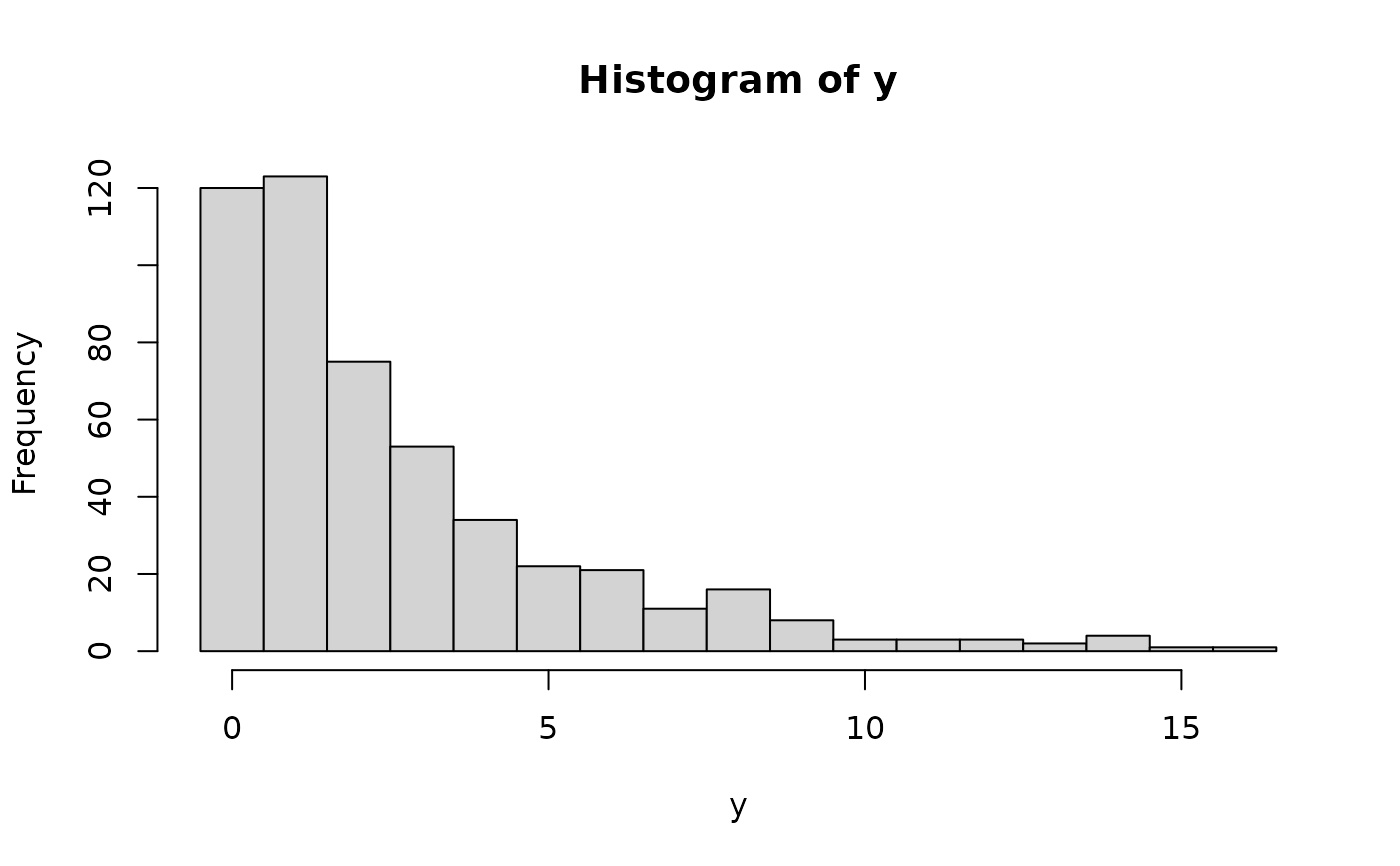
 ## corresponding empirical frequencies from a simulated sample
set.seed(0)
y <- rhnbinom(500, mu = 2.5, theta = 1, pi = 0.75)
hist(y, breaks = -1:max(y) + 0.5)
## corresponding empirical frequencies from a simulated sample
set.seed(0)
y <- rhnbinom(500, mu = 2.5, theta = 1, pi = 0.75)
hist(y, breaks = -1:max(y) + 0.5)